Yesterday we released version 0.5 of NIMBLE on our web site, r-nimble.org. (We’ll get it onto CRAN soon, but it has some special needs to work out.) NIMBLE tries to fill a gap in what R programmers and analysts can do with general hierarchical models. Packages like WinBUGS, OpenBUGS, JAGS and Stan provide a language for writing a model flexibly, and then they provide one flavor of MCMC. These have been workhorses of the Bayesian revolution, but they don’t provide much control over how the MCMC works (what samplers are used) or let one do anything else with the model (though Stan provides some additional fitting methods).
The idea of NIMBLE has been to provide a layer of programmability for algorithms that use models written in BUGS. We adopted BUGS as a model declaration language because these is so much BUGS code out there and so many books that use BUGS for teaching Bayesian statistics. Our implementation processes BUGS code in R and creates a model object that you can program with. For MCMC, we provide a default set of samplers, but these choices can be modified. It is easy to write your own sampler and add it to the MCMC. And it is easy to add new distributions and functions for use in BUGS code, something that hasn’t been possible (in any easy way) before. These features can allow big gains in MCMC efficiency.
MCMCs are heavily computational, so NIMBLE includes a compiler that generates C++ specific to a model and algorithm (MCMC samplers or otherwise), compiles it, loads it into R and gives you an interface to it. To be able to compile an algorithm, you need to write it as a nimbleFunction rather than a regular R function. nimbleFunctions can interact with model objects, and they can use a subset of R for math and flow-control. Among other things, the NIMBLE compiler automatically generates code for the Eigen C++ linear algebra library and manages all the necessary interfaces.
Actually, NIMBLE is not specific to MCMC or to Bayesian methods. You can write other algorithms to use whatever model you write in BUGS code. Here’s one simple example: in the past if you wanted to do a simulation study for a model written in BUGS code, you had to re-write the model in R just to simulate from it. With NIMBLE you can simulate from the model as written in BUGS and have complete control over what parts of the model you use. You can also query the model about how nodes are related so that you can make an algorithm adapt to what it finds in a model. We have a set of sequential Monte Carlo (particle filter) methods in development that we’ll release soon. But the idea is that NIMBLE provides a platform for others to develop and disseminate model-generic algorithms.
NIMBLE also extends BUGS in a bunch of ways that I won’t go into here. And it has one major limitation right now: it doesn’t handle models with stochastic indices, like latent class membership models.
Here is a toy example of what it looks like to set up and run an MCMC using NIMBLE.
library(nimble) myBUGScode <- nimbleCode({ mu ~ dnorm(0, sd = 100) ## uninformative prior sigma ~ dunif(0, 100) for(i in 1:10) y[i] ~ dnorm(mu, sd = sigma) }) myModel <- nimbleModel(myBUGScode)
myData <- rnorm(10, mean = 2, sd = 5) myModel$setData(list(y = myData)) myModel$setInits(list(mu = 0, sigma = 1)) myMCMC <- buildMCMC(myModel) compiled <- compileNimble(myModel, myMCMC) compiled$myMCMC$run(10000)
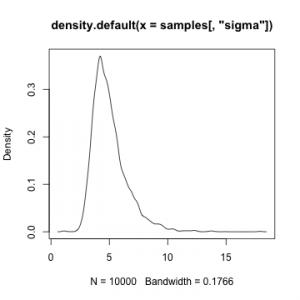
samples <- as.matrix(compiled$myMCMC$mvSamples) plot(density(samples[,'mu']))
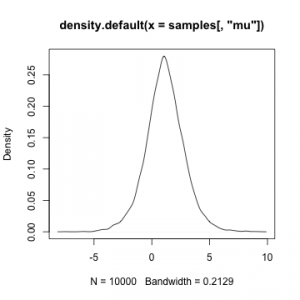
plot(density(samples[,'sigma']))