Version 0.9.1 of NIMBLE released
We’ve released the newest version of NIMBLE on CRAN and on our website. NIMBLE is a system for building and sharing analysis methods for statistical models, especially for hierarchical models and computationally-intensive methods (such as MCMC and SMC). Version 0.9.1 is primarily a bug fix release but also provides some minor improvements in functionality.
Users of NIMBLE in R 4.0 on Windows MUST upgrade to this release for NIMBLE to work.
New features and bug fixes include:
- switched to use of system2() from system() to avoid an issue on Windows in R 4.0;
- modified various adaptive MCMC samplers so the exponent controlling the scale decay of the adaptation is adjustable by user;
- allowed pmin() and pmax() to be used in models;
- improved handling of NA values in the dCRP distribution; and
- improved handling of cases where indexing goes beyond the extent of a variable in expandNodeNames() and related queries of model structure.
Please see the release notes on our website for more details.
nimbleEcology: custom NIMBLE distributions for ecologists
Prepared by Ben Goldstein.
What is nimbleEcology?
nimbleEcology is an auxiliary nimble package for ecologists.
nimbleEcology contains a set of distributions corresponding to some common ecological models. When the package is loaded, these distributions are registered to NIMBLE and can be used directly in models.
nimbleEcology contains distributions often used in modeling abundance, occupancy and capture-recapture studies.
Why use nimbleEcology?
Ecological models for abundance, occupancy and capture-recapture often involve many discrete latent states. Writing such models can be error-prone and in some cases can lead to slow MCMC mixing. We’ve put together a collection of distributions in nimble to make writing these models easier
- Easy to use. Using a nimbleEcology distribution is easier than writing out probabilities or hierarchical model descriptions.
- Minimize errors. You don’t have to lose hours looking for the misplaced minus sign; the distributions are checked and tested.
- Integrate over latent states. nimbleEcology implementations integrate or sum likelihoods over latent states. This eliminates the need for sampling these latent variables, which in some cases can provide efficiency gains, and allows maximum likelihood (ML) estimation methods with hierarchical models.
How to use
nimbleEcology can be installed directly from CRAN as follows.
install.packages("nimbleEcology")
Once nimbleEcology is installed, load it using library. It will also load nimble.
library(nimbleEcology)
## Loading required package: nimble
## nimble version 0.10.0 is loaded. ## For more information on NIMBLE and a User Manual, ## please visit http://R-nimble.org.
## ## Attaching package: 'nimble'
## The following object is masked from 'package:stats': ## ## simulate
## Loading nimbleEcology. ## Registering the following user-defined functions: ## dOcc, dDynOcc, dCJS, dHMM, dDHMM
Note the message indicating which distribution families have been loaded.
Which distributions are available?
The following distributions are available in nimbleEcology.
- dOcc (occupancy model)
- dDynOcc (dynamic occupancy model)
- dHMM (hidden Markov model)
- dDHMM (dynamic hidden Markov model)
- dCJS (Cormack-Jolly-Seber or mark-recapture model)
- dNmixture (N-mixture model)
- dYourNewDistribution Do you have a custom distribution that would fit the package? Are we missing a distribution you need? Let us know! We actively encourage contributions through GitHub or direct communication.
Example code
The following code illustrates a NIMBLE model definition for an occupancy model using nimbleEcology. The model is specified, built, and used to simulate some data according to the occupancy distribution.
library(nimbleEcology) occ_code <- nimbleCode({ psi ~ dunif(0, 1) p ~ dunif(0, 1) for (s in 1:nsite) { x[s, 1:nvisit] ~ dOcc_s(probOcc = psi, probDetect = p, len = nvisit) } }) occ_model <- nimbleModel(occ_code, constants = list(nsite = 10, nvisit = 5), inits = list(psi = 0.5, p = 0.5))
## defining model...
## building model...
## setting data and initial values...
## running calculate on model (any error reports that follow may simply reflect missing values in model variables) ... ## checking model sizes and dimensions... This model is not fully initialized. This is not an error. To see which variables are not initialized, use model$initializeInfo(). For more information on model initialization, see help(modelInitialization). ## model building finished.
set.seed(94) occ_model$simulate("x") occ_model$x
## [,1] [,2] [,3] [,4] [,5] ## [1,] 0 0 0 0 0 ## [2,] 0 0 0 0 0 ## [3,] 0 0 0 0 0 ## [4,] 0 0 0 0 0 ## [5,] 1 1 1 0 1 ## [6,] 0 0 0 1 0 ## [7,] 0 0 0 0 0 ## [8,] 0 0 0 0 1 ## [9,] 1 1 1 0 0 ## [10,] 0 1 0 0 0
How to learn more
Once the package is installed, you can check out the package vignette with vignette(“nimbleEcology”).
Documentation is available for each distribution family using the R syntax ?distribution, for example
?dHMM
For more detail on marginalization in these distributions, see the paper “One size does not fit all: Customizing MCMC methods for hierarchical models using NIMBLE” (Ponisio et al. 2020).
registration open for online NIMBLE short course, June 3-5, 2020
Registration is now open for a three-day online training workshop on NIMBLE, June 3-5, 2020, 9 am – 2 pm California time (noon – 5 pm US EDT). This online workshop will be held in place of our previously planned in-person workshop.
NIMBLE is a system for building and sharing analysis methods for statistical models, especially for hierarchical models and computationally-intensive methods (such as MCMC and SMC).
The workshop will cover:
- the basic concepts and workflows for using NIMBLE and converting BUGS or JAGS models to work in NIMBLE.
- overview of different MCMC sampling strategies and how to use them in NIMBLE.
- writing new distributions and functions for more flexible modeling and more efficient computation.
- tips and tricks for improving computational efficiency.
- using advanced model components, including Bayesian non-parametric distributions (based on Dirichlet process priors), conditional auto-regressive (CAR) models for spatially correlated random fields, and reversible jump samplers for variable selection.
- an introduction to programming new algorithms in NIMBLE.
- calling R and compiled C++ code from compiled NIMBLE models or functions.
If participant interests vary sufficiently, part of the third day will be split into two tracks. One of these will likely focus on ecological models. The other will be chosen based on attendee interest from topics such as (a) advanced NIMBLE programming including writing new MCMC samplers, (b) advanced spatial or Bayesian non-parametric modeling, or (c) non-MCMC algorithms in NIMBLE such as sequential Monte Carlo. Prior to the workshop, we will survey attendee interests and adjust content to meet attendee interests.
To register, please go here: https://na.eventscloud.com/540175. Registration is $120 (regular) or $60 (grad student).
registration open for NIMBLE short course, June 3-4, 2020 at UC Berkeley
Registration is now open for a two-day training workshop on NIMBLE, June 3-4, 2020 in Berkeley, California. NIMBLE is a system for building and sharing analysis methods for statistical models, especially for hierarchical models and computationally-intensive methods (such as MCMC and SMC).
The workshop will cover:
- the basic concepts and workflows for using NIMBLE and converting BUGS or JAGS models to work in NIMBLE.
- overview of different MCMC sampling strategies and how to use them in NIMBLE.
- writing new distributions and functions for more flexible modeling and more efficient computation.
- tips and tricks for improving computational efficiency.
- using advanced model components, including Bayesian non-parametric distributions (based on Dirichlet process priors), conditional auto-regressive (CAR) models for spatially correlated random fields, and reversible jump samplers for variable selection.
- an introduction to programming new algorithms in NIMBLE.
- calling R and compiled C++ code from compiled NIMBLE models or functions.
If participant interests vary sufficiently, the second half-day will be split into two tracks. One of these will likely focus on ecological models. The other will be chosen based on attendee interest from topics such as (a) advanced NIMBLE programming including writing new MCMC samplers, (b) advanced spatial or Bayesian non-parametric modeling, or (c) non-MCMC algorithms in NIMBLE such as sequential Monte Carlo. Prior to the workshop, we will survey attendee interests and adjust content to meet attendee interests.
To register, please go here: https://na.eventscloud.com/528790. Registration is $230 (regular) or $115 (student).
We have also reserved a block of rooms on campus for $137 per night, including breakfast and lunch. To reserve a room, go here: https://na.eventscloud.com/528792.
NIMBLE short course at March ENAR meeting in Nashville
We’ll be holding a half-day short course on NIMBLE on March 22 at the ENAR meeting in Nashville, Tennessee.
The annual ENAR meeting is a major biostatistics conference, sponsored by the eastern North American region of the International Biometric Society.
The short course will focus on usage of NIMBLE in applied statistics and is titled:
“Programming with hierarchical statistical models:
Using the BUGS-compatible NIMBLE system for MCMC and more”
More details on the conference and the short course are available at the conference website.
Variable selection in NIMBLE using reversible jump MCMC
Prepared by Sally Paganin.
Reversible Jump MCMC Overview
Reversible Jump MCMC (RJMCMC) is a general framework for MCMC simulation in which the dimension of the parameter space (i.e., the number of parameters) can vary between iterations of the Markov chain. It can be viewed as an extension of the Metropolis-Hastings algorithm onto more general state spaces. A common use case for RJMCMC is for variable selection in regression-style problems, where the dimension of the parameter space varies as variables are included or excluded from the regression specification.
Recently we added an RJMCMC sampler for variable selection to NIMBLE, using a univariate normal distribution as proposal distribution. There are two ways to use RJMCMC variable selection in your model. If you know the prior probability for inclusion of a variable in the model, you can use that directly in the RJMCMC without modifying your model. If you need the prior probability for inclusion in the model to be a model node itself, such as if it will have a prior and be estimated, you will need to write the model with indicator variables. In this post we will illustrate the basic usage of NIMBLE RJMCMC in both situations.
More information can be found in the NIMBLE User Manual and via help(configureRJ).
Linear regression example
In the following we consider a linear regression example in which we have 15 explanatory variables, and five of those are real effects while the others have no effect. First we simulate some data.
N <- 100 p <- 15 set.seed(1) X <- matrix(rnorm(N*p), nrow = N, ncol = p) true_betas <- c(c(0.1, 0.2, 0.3, 0.4, 0.5), rep(0, 10)) y <- rnorm(N, X%*%true_betas, sd = 1) summary(lm(y ~ X))
## ## Call: ## lm(formula = y ~ X) ## ## Residuals: ## Min 1Q Median 3Q Max ## -2.64569 -0.50496 0.01613 0.59480 1.97766 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) -0.04190 0.11511 -0.364 0.7168 ## X1 0.06574 0.12113 0.543 0.5888 ## X2 0.21162 0.11469 1.845 0.0686 . ## X3 0.16606 0.10823 1.534 0.1287 ## X4 0.66582 0.11376 5.853 9.06e-08 *** ## X5 0.51343 0.09351 5.490 4.18e-07 *** ## X6 0.01506 0.11399 0.132 0.8952 ## X7 -0.12203 0.10127 -1.205 0.2316 ## X8 0.18177 0.10168 1.788 0.0774 . ## X9 -0.09645 0.10617 -0.908 0.3663 ## X10 0.15986 0.11294 1.416 0.1606 ## X11 0.03806 0.10530 0.361 0.7186 ## X12 0.05354 0.10834 0.494 0.6225 ## X13 -0.02510 0.10516 -0.239 0.8119 ## X14 -0.07184 0.12842 -0.559 0.5774 ## X15 -0.04327 0.11236 -0.385 0.7011 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 1.035 on 84 degrees of freedom ## Multiple R-squared: 0.5337, Adjusted R-squared: 0.4505 ## F-statistic: 6.41 on 15 and 84 DF, p-value: 7.726e-09
Reversible jump with indicator variables
Next we set up the model. In this case we explicitly include indicator variables that include or exclude the corresponding predictor variable. For this example we assume the indicator variables are exchangeable and we include the inclusion probability in the inference.
lmIndicatorCode <- nimbleCode({ sigma ~ dunif(0, 20) ## uniform prior per Gelman (2006) psi ~ dunif(0,1) ## prior on inclusion probability for(i in 1:numVars) { z[i] ~ dbern(psi) ## indicator variable for each coefficient beta[i] ~ dnorm(0, sd = 100) zbeta[i] <- z[i] * beta[i] ## indicator * beta } for(i in 1:N) { pred.y[i] <- inprod(X[i, 1:numVars], zbeta[1:numVars]) y[i] ~ dnorm(pred.y[i], sd = sigma) } }) ## Set up the model. lmIndicatorConstants <- list(N = 100, numVars = 15) lmIndicatorInits <- list(sigma = 1, psi = 0.5, beta = rnorm(lmIndicatorConstants$numVars), z = sample(0:1, lmIndicatorConstants$numVars, 0.5)) lmIndicatorData <- list(y = y, X = X) lmIndicatorModel <- nimbleModel(code = lmIndicatorCode, constants = lmIndicatorConstants, inits = lmIndicatorInits, data = lmIndicatorData)
## defining model...
## building model...
## setting data and initial values...
## running calculate on model (any error reports that follow may simply reflect missing values in model variables) ... ## checking model sizes and dimensions... ## model building finished.
The above model code can potentially be used to set up variable selection in NIMBLE without using RJMCMC, since the indicator variables can turn the regression parameters off and on. However, in that case the MCMC sampling can be inefficient because a given regression parameter can wander widely in the parameter space when the corresponding variable is not in the model. This can make it difficult for the variable to be put back into the model, unless the prior for that variable is (perhaps artificially) made somewhat informative. Configuring RJMCMC sampling via our NIMBLE function configureRJ results in the MCMC not sampling the regression coefficients for variables for iterations where the variables are not in the model.
Configuring RJMCMC
The RJMCMC sampler can be added to the MCMC configuration by calling the function configureRJ(). In the example considered we introduced z as indicator variables associated with the regression coefficients beta. We can pass these, respectively, to configureRJ using the arguments indicatorNodes and targetNodes. The control arguments allow one to specify the mean and the scale of the normal proposal distribution used when proposing to put a coefficient back into the model.
lmIndicatorConf <- configureMCMC(lmIndicatorModel) lmIndicatorConf$addMonitors('z')
## thin = 1: sigma, psi, beta, z
configureRJ(lmIndicatorConf, targetNodes = 'beta', indicatorNodes = 'z', control = list(mean = 0, scale = .2))
Checking the assigned samplers we see that the indicator variables are each assigned an RJ_indicator sampler whose targetNode is the corresponding coefficient, while the beta parameters have a RJ_toggled sampler. The latter sampler is a modified version of the original sampler to the targetNode that is invoked only when the variable is currently in the model.
## Check the assigned samplers lmIndicatorConf$printSamplers(c("z[1]", "beta[1]"))
## [3] RJ_indicator sampler: z[1], mean: 0, scale: 0.2, targetNode: beta[1] ## [4] RJ_toggled sampler: beta[1], samplerType: conjugate_dnorm_dnorm
Build and run the RJMCMC
mcmcIndicatorRJ <- buildMCMC(lmIndicatorConf) cIndicatorModel <- compileNimble(lmIndicatorModel)
## compiling... this may take a minute. Use 'showCompilerOutput = TRUE' to see C++ compilation details.
## compilation finished.
CMCMCIndicatorRJ <- compileNimble(mcmcIndicatorRJ, project = lmIndicatorModel)
## compiling... this may take a minute. Use 'showCompilerOutput = TRUE' to see C++ compilation details. ## compilation finished.
set.seed(1) system.time(samplesIndicator <- runMCMC(CMCMCIndicatorRJ, niter = 10000, nburnin = 1000))
## running chain 1...
## |-------------|-------------|-------------|-------------| ## |-------------------------------------------------------|
## user system elapsed ## 2.749 0.004 2.753
Looking at the results
We can look at the sampled values of the indicator and corresponding coefficient for some of the variables.
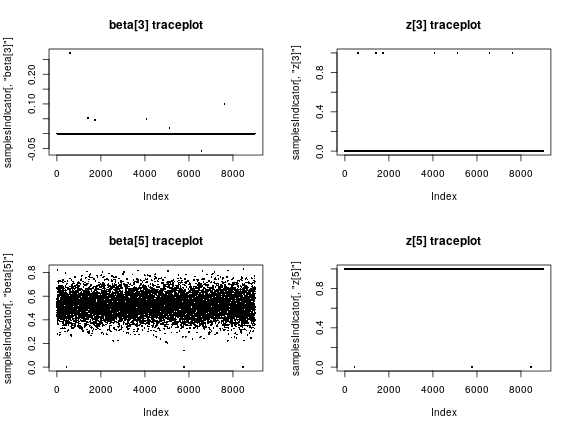
par(mfrow = c(2, 2)) plot(samplesIndicator[,'beta[3]'], pch = 16, cex = 0.4, main = "beta[3] traceplot") plot(samplesIndicator[,'z[3]'], pch = 16, cex = 0.4, main = "z[3] traceplot") plot(samplesIndicator[,'beta[5]'], pch = 16, cex = 0.4, main = "beta[5] traceplot") plot(samplesIndicator[,'z[5]'], pch = 16, cex = 0.4, main = "z[5] traceplot")
Individual inclusion probabilities
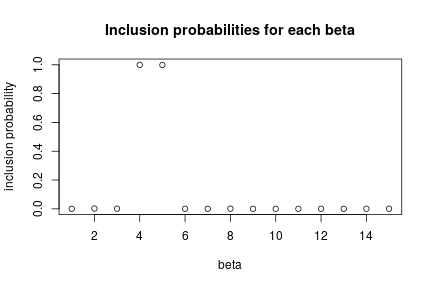
Now let’s look at the inference on the variable selection problem. We see that the fourth and fifth predictors are almost always included (these are the ones with the largest true coefficient values), while the others, including some variables that are truly associated with the outcome but have smaller true coefficient values, are almost never included.
par(mfrow = c(1, 1)) zCols <- grep("z\\[", colnames(samplesIndicator)) posterior_inclusion_prob <- colMeans(samplesIndicator[, zCols]) plot(1:length(true_betas), posterior_inclusion_prob, xlab = "beta", ylab = "inclusion probability", main = "Inclusion probabilities for each beta")
Reversible jump without indicator variables
If we assume that the inclusion probabilities for the coefficients are known, we can use the RJMCMC with model code written without indicator variables.
lmNoIndicatorCode <- nimbleCode({ sigma ~ dunif(0, 20) for(i in 1:numVars) { beta[i] ~ dnorm(0, sd = 100) } for(i in 1:N) { pred.y[i] <- inprod(X[i, 1:numVars], beta[1:numVars]) y[i] ~ dnorm(pred.y[i], sd = sigma) } }) ## Define model constants, inits and data lmNoIndicatorConstants <- list(N = 100, numVars = 15) lmNoIndicatorInits <- list(sigma = 1, beta = rnorm(lmNoIndicatorConstants$numVars)) lmNoIndicatorData <- list(y = y, X = X) lmNoIndicatorModel <- nimbleModel(code = lmNoIndicatorCode, constants = lmNoIndicatorConstants, inits = lmNoIndicatorInits, data = lmNoIndicatorData)
## defining model...
## building model...
## setting data and initial values...
## running calculate on model (any error reports that follow may simply reflect missing values in model variables) ... ## checking model sizes and dimensions... ## model building finished.
Configuring RJMCMC with no indicator variables
Again, the RJMCMC sampler can be added to the MCMC configuration by calling the function configureRJ() for nodes specified in targetNodes, but since there are no indicator variables we need to provide the prior inclusion probabilities. We use the priorProb argument, and we can provide either a vector of values or a common value.
lmNoIndicatorConf <- configureMCMC(lmNoIndicatorModel) configureRJ(lmNoIndicatorConf, targetNodes = 'beta', priorProb = 0.5, control = list(mean = 0, scale = .2))
Since there are no indicator variables in this case, a RJ_fixed_prior sampler is assigned directly to each of coefficents along with the RJ_toggled sampler, which still uses the default sampler for the node, but only if the corresponding variable is in the model at a given iteration. In addition in this case one can set the coefficient to a value different from zero via the fixedValue argument in the control list.
## Check the assigned samplers lmNoIndicatorConf$printSamplers(c("beta[1]"))
## [2] RJ_fixed_prior sampler: beta[1], priorProb: 0.5, mean: 0, scale: 0.2, fixedValue: 0 ## [3] RJ_toggled sampler: beta[1], samplerType: conjugate_dnorm_dnorm, fixedValue: 0
Build and run the RJMCMC
mcmcNoIndicatorRJ <- buildMCMC(lmNoIndicatorConf) cNoIndicatorModel <- compileNimble(lmNoIndicatorModel)
## compiling... this may take a minute. Use 'showCompilerOutput = TRUE' to see C++ compilation details.
## compilation finished.
CMCMCNoIndicatorRJ <- compileNimble(mcmcNoIndicatorRJ, project = lmNoIndicatorModel)
## compiling... this may take a minute. Use 'showCompilerOutput = TRUE' to see C++ compilation details. ## compilation finished.
set.seed(100) system.time(samplesNoIndicator <- runMCMC(CMCMCNoIndicatorRJ, niter = 10000, nburnin = 1000))
## running chain 1...
## |-------------|-------------|-------------|-------------| ## |-------------------------------------------------------|
## user system elapsed ## 3.014 0.004 3.050
Looking at the results
In this case we just look at one of the model coefficients.
plot(samplesNoIndicator[,'beta[1]'], pch = 16, cex = 0.4, main = "beta[1] traceplot")

Individual inclusion proportion
We can calculate the proportion of times each coefficient is included in the model.
betaCols <- grep("beta\\[", colnames(samplesNoIndicator)) posterior_inclusion_proportions <- colMeans(apply(samplesNoIndicator[, betaCols], 2, function(x) x != 0)) posterior_inclusion_proportions
## beta[1] beta[2] beta[3] beta[4] beta[5] ## 0.0017777778 0.0097777778 0.0017777778 1.0000000000 0.9996666667 ## beta[6] beta[7] beta[8] beta[9] beta[10] ## 0.0015555556 0.0015555556 0.0031111111 0.0018888889 0.0028888889 ## beta[11] beta[12] beta[13] beta[14] beta[15] ## 0.0006666667 0.0015555556 0.0007777778 0.0024444444 0.0007777778
Version 0.9.0 of NIMBLE released
We’ve released the newest version of NIMBLE on CRAN and on our website. NIMBLE is a system for building and sharing analysis methods for statistical models, especially for hierarchical models and computationally-intensive methods (such as MCMC and SMC). Version 0.9.0 provides some new features as well as providing a variety of speed improvements, better output handling, and bug fixes.
New features and bug fixes include:
- added an iterated filtering 2 (IF2) algorithm (a sequential Monte Carlo (SMC) method) for parameter estimation via maximum likelihood;
- fixed several bugs in our SMC algorithms;
- improved the speed of MCMC configuration;
- improved the user interface for interacting with the MCMC configuration; and
- improved our conjugacy checking system to detect some additional cases of conjugacy.
Please see the release notes on our website for more details.
New NIMBLE code examples
We’ve posted a variety of new code examples for NIMBLE at https://r-nimble.org/examples.
These include some introductory code for getting started with NIMBLE, as well as examples of setting up specific kinds of models such as GLMMs, spatial models, item-response theory models, and a variety of models widely used in ecology. We also have examples of doing variable selection via reversible jump MCMC, using MCEM, and writing new distributions for use with NIMBLE.
And if you have examples that you think might be useful to make available to others, please get in touch with us.
NIMBLE short course, June 3-4, 2020 at UC Berkeley
We’ll be holding a two-day training workshop on NIMBLE, June 3-4, 2020 in Berkeley, California. NIMBLE is a system for building and sharing analysis methods for statistical models, especially for hierarchical models and computationally-intensive methods (such as MCMC and SMC).
The tutorial will cover
- the basic concepts and workflows for using NIMBLE and converting BUGS or JAGS models to work in NIMBLE.
- overview of different MCMC sampling strategies and how to use them in NIMBLE.
- writing new distributions and functions for more flexible modeling and more efficient computation.
- tips and tricks for improving computational efficiency.
- using advanced model components, including Bayesian non-parametric distributions (based on Dirichlet process priors), conditional auto-regressive (CAR) models for spatially correlated random fields, and reversible jump samplers for variable selection.
- an introduction to programming new algorithms in NIMBLE.
- calling R and compiled C++ code from compiled NIMBLE models or functions.
If participant interests vary sufficiently, the second half-day will be split into two tracks. One of these will likely focus on ecological models. The other will be chosen based on attendee interest from topics such as (a) advanced NIMBLE programming including writing new MCMC samplers, (b) advanced spatial or Bayesian non-parametric modeling, or (c) non-MCMC algorithms in NIMBLE such as sequential Monte Carlo. Prior to the workshop, we will survey attendee interests and adjust content to meet attendee interests.
If you are interested in attending, please pre-register to hold a spot at https://forms.gle/6AtNgfdUdvhni32Q6. The form also asks if you are interested in a relatively cheap dormitory-style housing option. No payment is necessary to pre-register. Fees to finalize registration will be $230 (regular) or $115 (student). We hope to be able to offer student travel awards; more information will follow.
The workshop will assume attendees have a basic understanding of hierarchical/Bayesian models and MCMC, the BUGS (or JAGS) model language, and some familiarity with R.
NIMBLE short course at Bayes Comp 2020 conference
We’ll be giving a short course on NIMBLE on January 7, 2020 at the Bayes Comp 2020 conference being held January 7-10 in Gainesville, Florida, USA.
Bayes Comp is a popular biennial ISBA-sponsored conference focused on computational methods/algorithms/technologies for Bayesian inference.
The short course focuses on programming algorithms in NIMBLE and is titled:
“Developing, modifying, and sharing Bayesian algorithms (MCMC samplers, SMC, and more) using the NIMBLE platform in R.”
More details on the conference and the short course are available at the conference website.